Proton Transfer Analysis Toolkit#
ai2-kit algorithm proton-transfer
Introduction#
The proton transfer analysis toolkit is a set of commands and Python functions to analyze proton transfer events in a trajectory file.[YWL23]
Usage#
Trajectory Analysis#
ai2-kit algorithm proton-transfer analyze
This command will analyze the events of proton transfer in a trajectory and dump analysis results as files, which contains the coordinate of a proton indicator and the paths of proton transfer. The result will be used by the other commands in this toolkit later.
Options#
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
input_traj |
the trajectory file to analyses, currently only xyz format is supported. |
str |
(Required) |
|
out_dir |
the output directory to save analysis result |
str |
(Required) |
|
cell |
pbc parameters |
List[float] |
(Required) |
|
acceptor_elements |
the elements of atoms that can act as acceptor |
List[str] |
(Required) |
|
initial_donors |
indexes of donor atoms where proton transfer may be initiated |
List[int] |
(Required) |
|
core_num |
number of cpu cores used in the analysis |
int |
4 |
|
dt |
the time step between frames |
float |
0.0005 |
|
r_a |
search range of acceptor |
float |
4.0 |
|
r_h |
the range of searching proton |
float |
1.3 |
|
rho_0 |
the rate of the weights change |
float |
0.4545 |
|
rho_max |
the critical value of proton transfer |
float |
0.5 |
|
max_depth |
the maximum length of proton paths |
int |
4 |
|
g_threshold |
the threshold for determining whether to join a node to the proton transfer paths |
float |
0.0001 |
|
Examples#
You can use the data in the doc/res directory to run the following examples.
ai2-kit algorithm proton-transfer analyze \
--input_traj ./doc/res/proton-transfer-test-trajectory.xyz \
--out_dir ./result \
--cell "[12.740,13.399,40.985,90,90,90]" \
--acceptor_elements '["O"]' \
--initial_donors '[240,255,246]'
The output of the above command will be in a directory named result. Files in the directory are named after the index of initial donors, 240.jsonl for example. The content of a output files is like below:
[[6.568212710702677, 3.9114527694565866, 15.197925887324345], []]
[[6.568089962005615, 3.9143550395965576, 15.198609828948975], [[588, 241]]]
[[6.568215087380366, 4.006908148832614, 15.244050368181686], []]
The analysis result is not supposed to be read or modified by human. It will be used by the other commands in this toolkit later.
Visualization#
ai2-kit algorithm proton-transfer visualize
This command will visualize the proton transfer events in a trajectory by adding the proton indicators and marking the positions of donors in each frame base on the analysis result.
Options#
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
analysis_result |
the directory of analysis result |
str |
(Required) |
|
input_traj |
the original trajectory file |
str |
(Required) |
|
output_traj |
the processed trajectory file with labeled |
str |
(Required) |
|
initial_donor |
the index of a initial donor |
int |
(Required) |
|
cell |
pbc parameters |
List[float] |
(Required) |
|
Examples#
ai2-kit algorithm proton-transfer visualize \
--analysis_result ./result \
--input_traj ./doc/res/proton-transfer-test-trajectory.xyz \
--output_traj ./labeled-traj.xyz \
--initial_donor 240 \
--cell '[12.745,13.399,40.985,90,90,90]'
Now we can use tools such as ASE or VMD to visualize the output trajectory.
https://github.com/chenggroup/ai2-kit/assets/3314130/a973cd0e-044f-405a-8476-ee37a9b4d1b7
Show Transfers Paths#
ai2-kit algorithm proton-transfer show-transfer-paths
This command will show proton transfer paths in a human readable format.(This command will also dump proton infomations as files, which contains the index of proton and the time of proton transfer. The result will be used by the other commands in this toolkit later.)
Options#
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
analysis_result |
the directory of analysis result |
str |
(Required) |
|
initial_donor |
the index of a initial donor |
int |
(Required) |
|
Examples#
ai2-kit algorithm proton-transfer show-transfer-paths \
--analysis_result ./result \
--initial_donor 255
The output of the above command is shown below:
transfer_paths
transfer_path_index Snapshot
255(257)->258 5815
258(257)->255 5827
255(257)->477 13742
477(257)->255 13773
255(257)->477 13787
477(257)->255 13802
255(257)->477 13814
477(478)->49 13818
... ...
Show Type Change#
ai2-kit algorithm proton-transfer show-type-change
Options#
This command will show type changes within proton transfer events.
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
analysis_result |
the directory of analysis result |
str |
(Required) |
|
atom_types |
different types of atoms |
dict |
(Required) |
|
donors |
the donors that we want to analyze |
List[int] |
(Required) |
|
Examples#
ai2-kit algorithm proton-transfer show-type-change \
--analysis_result ./result \
--atom_type '{"Bridge_O":[169,139,229,199,49,19,109,79,40,10,70,100,160,130,220,190],"Water_O":[258,255,252,261,64,240,249,246,279,276,285,282,267,264,273,270]}' \
--donors '[240,255]'
The output of the above command will be like below:
proton transfer type change
-------------------------------------
Path_index start_Snapshot end_Snapshot
Bridge_O<->Bridge_O
49 -> 477 -> 49 13818 17738
19 -> 630 -> 19 43212 44114
49 -> 477 -> 630 -> 19 42875 43212
Bridge_O<->Water_O
255 -> 477 -> 49 13802 13818
49 -> 477 -> 255 17738 19430
Water_O<->Water_O
255 -> 258 0 5815
258 -> 255 5815 5827
240 -> 588 -> 240 0 9034
240 -> 306 -> 240 9034 35042
255 -> 477 -> 255 5827 13773
Calculate Distances#
ai2-kit algorithm proton-transfer calculate-distances
This command will calculate the distance from the proton to the nearest interface and dump results as files.
Options#
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
analysis_result |
the directory of analysis result |
str |
(Required) |
|
input_traj |
the trajectory file to analyses, currently only xyz format is supported. |
str |
(Required) |
|
upper_index |
upper interface atomic index |
List[int] |
(Required) |
|
lower_index |
lower interface atomic index |
List[int] |
(Required) |
|
initial_donor |
the index of a initial donor |
int |
(Required) |
|
interval |
each time interval the number of frames |
int |
1 |
|
Examples#
ai2-kit algorithm proton-transfer calculate-distances \
--analysis_result ./result \
--input_traj ./test.xyz \
--upper_index '[1,2]' \
--lower_index '[3,4]' \
--initial_donor 255 \
--interval 1
The content of the output file will be like below:
4.10318
4.11708
4.131689999999999
4.14673
4.16193
4.177009999999999
4.1916899999999995
4.20575
4.218959999999999
4.231159999999999
4.24221
4.252049999999999
4.260639999999999
4.26798
4.274089999999999
4.279
...
Show Distance Change#
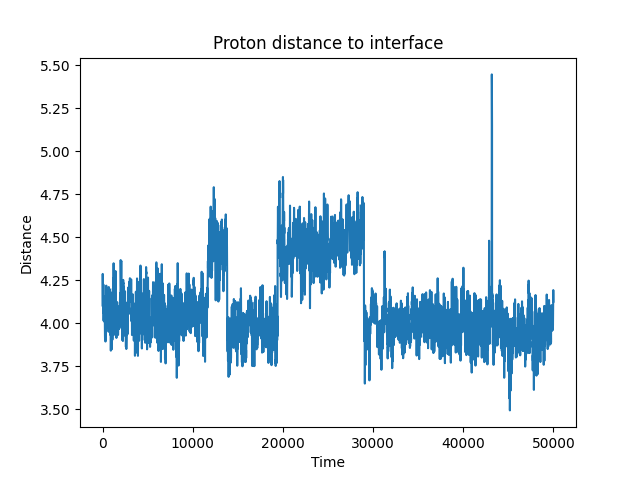
ai2-kit algorithm proton-transfer show-distance-change
This command will draw the distance change over time.
Options#
Option |
Description |
Type |
Default |
Example |
|---|---|---|---|---|
analysis_result |
the directory of analysis result |
str |
(Required) |
|
initial_donor |
the index of a initial donor |
int |
(Required) |
|
Examples#
ai2-kit algorithm proton-transfer show-distance-change \
--analysis_result ./result \
--initial_donor 255
The output of the above command will be like below: